September 16, 2024
by UCLA Engineering Institute for Technology Advancement
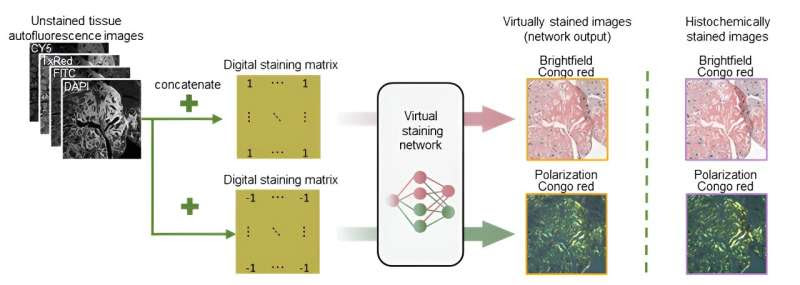
Virtual birefringence imaging and histological staining of amyloid deposits in label-free tissue. Virtual staining network and digital staining matrix framework generate two output images (brightfield and birefringence image channels), eliminating the need for polarization imaging and traditional chemical stains like Congo red used in clinical workflow. Credit: UCLA Engineering Institute for Technology Advancement
Researchers at the University of California, Los Angeles (UCLA) have pioneered a groundbreaking approach in the imaging and detection of amyloid deposits in tissue samples. The innovative method leverages deep learning and autofluorescence microscopy to achieve virtual birefringence imaging and histological staining, eliminating the need for polarization imaging and traditional chemical stains like Congo red.
Systemic amyloidosis, a condition characterized by the accumulation of misfolded proteins in organs and tissues, poses significant diagnostic challenges. Amyloidosis affects several million people every year, often leading to severe organ damage, heart failure, and high mortality rates if not diagnosed and treated early.
Traditionally, Congo red staining under polarized light microscopy has been the gold standard for visualizing amyloid deposits. However, this method is labor-intensive, costly, and subject to variability that can lead to false diagnoses.
The new technique, detailed in Nature Communications, utilizes a single neural network to transform autofluorescence images of label-free tissue into high-fidelity brightfield and polarized microscopy images that mirror those obtained through traditional histochemical staining and polarization microscopy.
The technique was tested on cardiac tissue samples, showing that virtually stained images provided consistent and reliable identification of amyloid patterns compared to traditional methods, also eliminating the need for chemical staining and specialized polarization microscopes, potentially speeding up diagnosis and reducing costs.
This virtual staining process not only matches but, in some cases, surpasses the quality of conventional methods, as validated by multiple board-certified pathologists from UCLA as well as USC and the Hadassah Hebrew University Medical Center.
Dr. Aydogan Ozcan, the senior author of the study and the Volgenau Chair for Engineering Innovation at UCLA, explains, “Our deep learning model can perform both autofluorescence-to-birefringence and autofluorescence-to-brightfield image transformations, offering a reliable, consistent, and cost-effective alternative to traditional histology methods.
“This breakthrough could greatly enhance the speed and accuracy of amyloidosis diagnosis, reducing the risk of false negatives and improving patient outcomes.”
The study’s findings suggest that this virtual staining approach could be seamlessly integrated into existing clinical workflows, facilitating the broader adoption of digital pathology. The method requires no specialized optical components and can be implemented on standard digital pathology scanners, making it accessible to a wide range of health care settings.
“This innovation represents a significant step forward in the field of amyloidosis pathology,” said Dr. Ozcan. “It not only simplifies the diagnostic process but also holds potential for expanding the use of digital pathology in routine clinical practice, particularly in resource-limited settings.”
The researchers plan to expand their evaluations to other tissue types, such as kidney, liver, and spleen, to further validate the model’s clinical utility across different amyloidosis manifestations. They also aim to explore the development of automated detection systems to assist pathologists in identifying problematic areas, potentially improving diagnostic accuracy and reducing false negatives.
More information: Xilin Yang et al, Virtual birefringence imaging and histological staining of amyloid deposits in label-free tissue using autofluorescence microscopy and deep learning, Nature Communications (2024). DOI: 10.1038/s41467-024-52263-z
Journal information: Nature Communications
Provided by UCLA Engineering Institute for Technology Advancement

Leave a Reply