by Indiana University
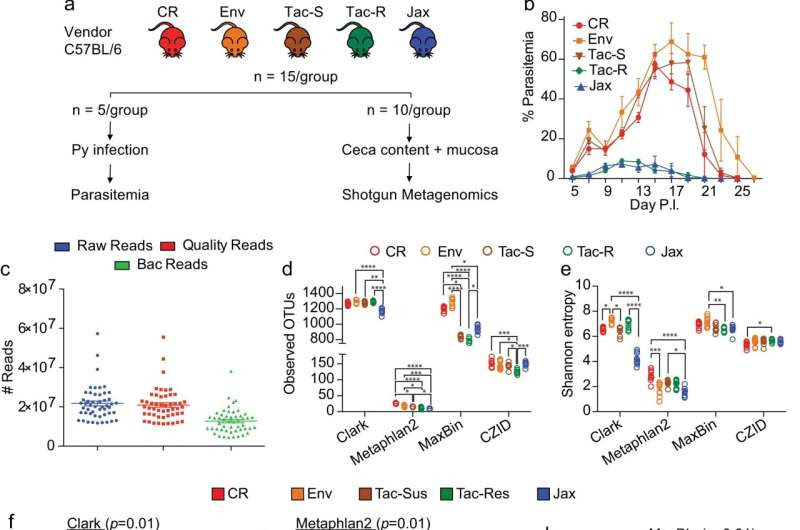
Shotgun metagenomics revealed distinct gut microbiota composition and genetic potential within and between the hyperparasitemia resistant and susceptible mice. a C57BL/6 mice were acquired from four different vendors (N = 15/group): Charles River Laboratories (CR), Envigo (Env), Taconic Biosciences (Tac), and Jackson Laboratory (Jax). Mice from Taconic Biosciences were obtained from two different facilities with differential susceptibility to Py hyperparasitemia12. Five mice from each group were infected with Py while the remaining ten mice were sacrificed to collect ceca content along with mucosa scrapes for shotgun metagenomics. b Parasitemia curve of mice infected with Py. c Number of fastq reads after shotgun sequencing and quality control. d Alpha diversity measured using observed taxonomic units (OTUs) defined at species level. e Alpha diversity measure by Shannon entropy. f Beta diversity shown by Principal coordinate analysis (PCoA) plot using Clark output at species level with Bray-Curtis distance. g Beta diversity shown by PCoA plot using Metaphlan2 output at species level with Bray-Curtis distance. h Beta diversity shown by PCoA plot using MaxBin output at species level with Bray-Curtis distance. i Beta diversity shown by PCoA plot using CZID output at bin level with Bray-Curtis distance. j Number of unique genes detected at 95% sequence homology. All data are mean ± SE (standard error) unless explicitly stated. Bacterial diversity was performed on normalized data. Mice resistant to hyperparasitemia are encircled (f–i). Alpha diversity significance were calculated with Kruskal Wallis test and beta diversity significance by pairwise Permutational multivariate analysis of variance (PERMANOVA). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Exact p values are shown in Source Data file. Py: Plasmodium yoelii 17XNL; Tac-R and Tac-S: Taconic mice resistant and susceptible to hyperparasitemia to Py infection respectively. Source data are provided in the Source Data file. Credit: Nature Communications (2023). DOI: 10.1038/s41467-023-42235-0
Indiana University School of Medicine researchers have identified multiple species of bacteria that, when present in the gut, are linked to an increased risk of developing severe malaria in humans and mice. Their findings, recently published in Nature Communications, could lead to the development of new approaches targeting gut bacteria to prevent severe malaria and associated deaths.
Malaria is a life-threatening infectious disease caused by parasites transmitted through the bite of infected mosquitoes. According to the World Health Organization’s latest World Malaria Report, an estimated 619,000 people died from malaria globally in 2021, with 76% of those deaths occurring in children age 5 or younger.
IU School of Medicine’s Nathan Schmidt, Ph.D., an associate professor of pediatrics with the Ryan White Center for Pediatric Infectious Disease and Global Health and the Herman B Wells Center for Pediatric Research, said previous efforts to combat the disease have led to several advancements in malaria treatment and prevention, including new vaccines and antimalarial drugs, insecticides to manage mosquito populations and improved health care processes.
However, he said new developments are desperately needed because the gains made in decreasing malaria-related deaths between the early 2000s and late 2010s have plateaued over the last five years.
“This plateau highlights the need for novel approaches to prevent malaria-related fatalities,” said Schmidt, whose research lab is focused on investigating this global health crisis and its critical impact on children. “Presently, there are no approaches that target gut microbiota. Therefore, we believe that our approach represents an exciting opportunity.”
In a pivotal 2016 article published in PNAS, Schmidt and his colleagues made a groundbreaking discovery in their experimental models: the gut microbiota has the capability to influence the severity of malaria. This revelation ignited their determination to pinpoint the precise microorganisms, called “Bacteroides,” within the intestinal tract that orchestrate this effect.
In their latest study, the researchers found mice harboring particular species of Bacteroides were notably associated with an elevated risk of severe malaria. A similar correlation was also observed in the intestinal tracts of children afflicted with severe malaria.
Most of the Schmidt lab’s research has been conducted using mouse models of malaria. Thanks to collaboration with several colleagues in the field, the research team was able to extend its observations by studying approximately 50 children with malaria in Uganda. They plan to continue their clinical observations by working with a cohort of over 500 children with malaria.
This collaboration was made possible by the joint efforts of Chandy John, MD, MS, of IU School of Medicine; Ruth Namazzi, MB ChB, MMEd, of Makerere University; and Robert Opoka, MD, MPH, of Global Health Uganda. Together, they are evaluating how severe malaria may affect child neurodevelopment by studying children from households with a history of severe malaria.
While these children may not display any symptoms of illness, some carry the malaria parasite in their blood, allowing researchers to explore risk factors associated with the development of severe malaria, including variations observed in the microbiome.
“Dr. Namazzi, Dr. Opoka and I aren’t experts in the microbiome, so we collaborated with Nathan [Schmidt] on this part of the study since he is an expert,” said John, who is the Ryan White Professor of Pediatrics at IU School of Medicine. “I believe Nathan’s findings are important because they point to the possibility that certain bacteria or combinations of bacteria in the gut may predispose a child to severe malaria. This opens the way to thinking about how we might alter those combinations in the gut to try to protect children from severe malaria.”
In addition to studying the expanded cohort in Uganda, Schmidt and his team will also collaborate with researchers in Malawi and Mali to get a broader sense of trends present between gut microbiota and malaria across Africa.
“Beyond our efforts to assess the contribution of gut bacteria towards severe malaria in diverse African populations, we have initiated pre-clinical efforts to target gut bacteria that cause susceptibility to severe malaria,” Schmidt said. “Our long-term aspiration is to move a treatment into the clinic.”
More information: Rabindra K. Mandal et al, Gut Bacteroides act in a microbial consortium to cause susceptibility to severe malaria, Nature Communications (2023). DOI: 10.1038/s41467-023-42235-0
Journal information: Proceedings of the National Academy of Sciences , Nature Communications
Provided by Indiana University

Leave a Reply