by University of Veterinary Medicine—Vienna
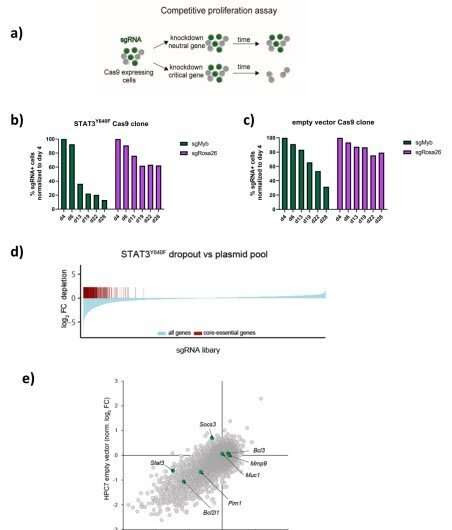
Genome-wide CRISPR/Cas9-based loss-of-function screen identifies genetic dependencies of STAT3Y640F-driven cells a) Schematic illustration of a competitive proliferation assay. b) Competitive proliferation assays of clonal Cas9-expressing STATY640F-driven HPC7 or c) clonal Cas9-HPC7 empty vector cells expressing IRF670 and sgRNAs targeting either Myb (positive control) or the Rosa26 locus (negative control). d) Waterfall plot of log2-fold-changebased gene knockout effects derived from sgRNA abundances in STAT3Y640F-driven HPC7 cells at the screen endpoint compared to the plasmid pool. Screen quality was assessed by the depletion of core-essential genes. e) Comparative analysis of two CRISPR-based LOF screens, canonical STAT3 target genes are highlighted in green.
A research team led by scientists from the University of Veterinary Medicine Vienna (Vetmeduni) has taken an important step toward a better understanding of certain difficult-to-treat blood cancers. The researchers have identified the SBNO2 protein as a critical mediator of certain types of leukemia—a finding that could potentially lead to novel therapeutic approaches.
In blood cancers such as leukemia, molecular signaling pathways are often deregulated. This leads to uncontrolled cell division. A frequently found cause of such a deregulation is a mutation in the gene STAT3, which is a focus of the Viennese research group. Current therapeutic options lead to an unsatisfactory response rate and no cure for these STAT3-driven malignancies exist. Therefore, the Vetmeduni researchers investigated the mechanism behind these blood cancers in a study that was recently published in the journal Blood.
But which genes are activated by mutant STAT3? And which of these genes are necessary for cancer cell survival? Veronika Sexl, supervisor of the study and head of the Institute of Pharmacology and Toxicology at Vetmeduni, said, “We know from previous research that STAT3 plays an important role in hematological cancers, but little is known about the exact molecular mechanisms contributing to disease development. As targeted therapy of mutated STAT3 in patients is currently not possible, our focus lies on finding new therapeutic approaches.”
Protein SBNO2 essential for leukemia driven by STAT3 mutations
In a collaborative study involving international experts in the field, the latest genomics technology was used to investigate mechanisms associated with mutated STAT3. The CRISPR-Cas9 screening technology, referred to as the “gene scissors” technique, helped to identify an essential protein in mutant STAT3-driven malignancies: SBNO2.
“The exciting thing about our work is that we discovered a global mechanism that applies to all STAT3-driven hematological malignancies,” says the study’s first author, Tania Brandstötter from the Institute of Pharmacology and Toxicology at Vetmeduni.
Knowledge about the importance of SBNO2 in STAT3-driven leukemias and lymphomas reveals novel therapeutic opportunities and provides the basis for a number of future studies.

Leave a Reply