by The Hospital for Sick Children
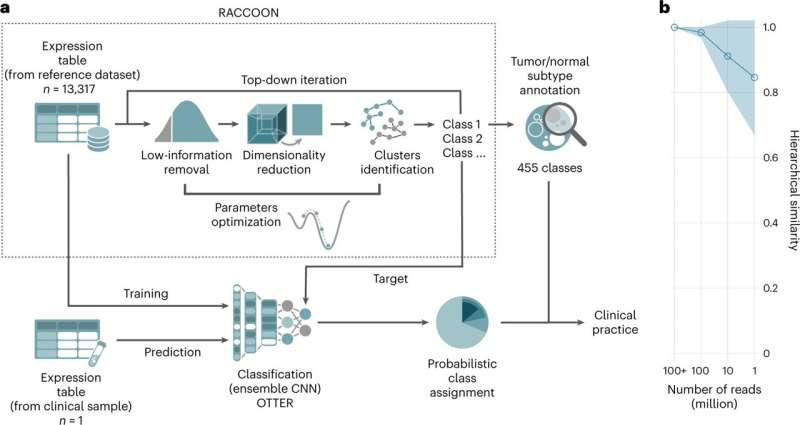
A platform for clustering and classification of RNA-seq data. a, Schematic representation of the steps involved in our RNA-seq tumor subtype identification protocol. We first built an extensive reference hierarchy of tumor and normal subtypes using RACCOON, a novel scale-adaptive clustering framework. This hierarchy was then used as a target for OTTER, an ensemble of CNN classifiers, which can be employed to identify multiple tumor and normal tissue components in samples from clinical practice. b, OTTER performance as a function of the number of sequenced reads. This is quantified as the hierarchical similarity (Methods) between the prediction probabilities obtained on subsampled data and the original sample (>1 × 108 reads). Values are presented as mean and standard deviation of six tumor samples with reads randomly subsampled five times each. Expression counts were obtained with a STAR + RSEM pipeline. Credit: Nature Medicine (2023). DOI: 10.1038/s41591-023-02221-x
In the first broad comparison of pediatric and adult cancer, researchers at The Hospital for Sick Children (SickKids) have analyzed 13,000 individual cancers and built an “atlas” of pediatric cancer using a novel machine-learning algorithm.
The diagnosis of cancer is, for an estimated 18.1 million people worldwide per year, mostly reliant on the microscopic examination and detection of specific proteins. The accuracy of these methods is variable, and improvements are not easily shared between institutes. This is especially true for pediatric cancer, which is the most frequent cause of death-by-disease in children past infancy in the developed world.
“As the burden of cancer increases worldwide, the complexity of cancer diagnostics is expected to grow unless new methods are developed,” explains Dr. Adam Shlien, a Senior Scientist in the Genetics & Genome Biology program whose team developed this algorithm. “Our platform can be used at any hospital to increase the speed and accuracy of diagnosing cancer, even for rare types.”
Analysis of transcriptome illuminates uniqueness of pediatric cancer
Described in a new study published in Nature Medicine, this machine-learning algorithm classifies every known major type of childhood cancer and can refine, or match, a given cancer diagnosis for 85% of pediatric cancer patients.
Unlike other tools for detection and diagnosis, such as a cancer panel test which looks for mutations in specific genes or other methods which may analyze the genome alone, this machine-learning algorithm analyzes a person’s entire transcriptome. While the genome is made up of all the DNA in a cell, only a portion of this genetic code is copied into RNA molecules, known as the transcriptome.
“Just because you have a very busy cancer genome, doesn’t mean that everything is being acted open,” says Dr. Federico Comitani, a Research Associate in the Genetics & Genome Biology program and first author on the study. “By analyzing the full transcriptome, we can find each tumor’s core features and collect a clearer picture of cancer activity specific to each individual.”
In addition to identifying significant differences between cancer types, the large amount of data collected by the research team and magnification provided by the platform allowed researchers to identify 455 subtypes of cancer. This large number of subtypes lends support to the idea that most childhood cancers share a common ancestry and then differentiate into a multitude of specific tumor subtypes.
“We were able to see, for the first time, subtle differences within cancer subtypes. Childhood cancers display more transcriptional variability—the number of the genes expressed in a cell—than adult cancers,” says Shlien, who holds a Canada Research Chair in Childhood Cancer Genomics and is an Associate Director in the Department of Pediatric Laboratory Medicine. “This gives us a radically new way to look at cancer and potentially identify the prognosis of cancers, and the possibility of changing our understanding of cancer.”
Classifier can improve diagnosis for pediatric cancers
The tool is already playing an important role in the faster and more accurate diagnosis of cancer as part of the SickKids Cancer Sequencing program (KiCS), which provides comprehensive genetic sequencing for children with cancer.
In cases of neuroblastoma, the most common extra-cranial solid tumor in children, the subtypes identified by this tool predicted significant differences in tumor differentiation and patient survival. Similarly, findings from the platform explained the inconsistent response of sarcomas, tumors of the bone and soft tissue, to immunotherapy by uncovering an imbalance of immune cells, informing potential new therapeutic approaches.
“As we add more samples to this growing atlas and validate it with even larger data sets and sample types, our classifier has the potential to become a universal test for diagnosing pediatric cancer,” says Shlien.
This RNA platform is currently being used on a research use only basis by a number of early adopter cancer centers worldwide, allowing physicians to compare their patient’s diagnosis to cancer types identified by the platform and receive a digital diagnosis. Work is also underway to bring this tool to the broader community as a platform to enable diagnostic testing and the acceleration of cancer drug product development.

Leave a Reply