by University of Michigan
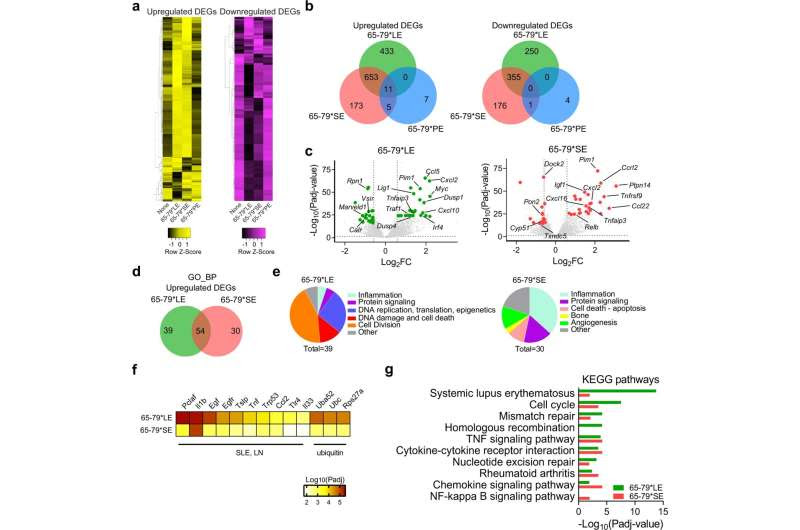
Transcriptional modulation by the LE (Model A: epitope + IFN-γ, versus IFN-γ alone). a Heatmaps showing unsupervised clustering of upregulated (left, yellow) and downregulated (right, purple) DEGs for 65-79*LE, 65-79*SE and 65-79*PE. DEGs were selected based on a > 3-fold change for 65-79*LE. All depicted DEGs had a Padj < 0.05. b Venn diagrams showing comparison of DEGs for 65-79*LE, 65-79*SE and 65-79*PE. c Volcano plots comparing transcriptional modulation by 65-79*LE and 65-79*SE. Green dots denote SLE-relevant genes modulated by 65-79*LE; red dots denote RA-relevant genes modulated by 65-79*SE. d Venn diagram comparing GO-BP terms enrichment among upregulated DEGs for 65-79*LE versus 65-79*SE. e Pie charts showing differential thematic clustering of unique GO-BP term enrichment among 65-79*LE- (left) versus 65-79*SE- (right) upregulated DEGs. f Heatmap of top-ranked SLE- or ubiquitination-associated URs that are predicted to be involved in 65-79*LE transcriptional modulation, as compared to their predicted involvement in 65-79*SE transcriptional modulation. g Notable KEGG pathway enrichment among 65-79*LE-upregulated DEGs, versus 65-79*SE. Data in (b–g) are based on DEGs with a fold change >1.5 and Padj < 0.05. All data are based on 6 biological replicates in 2 independent experiments. Credit: Communications Biology (2022). DOI: 10.1038/s42003-022-03717-x
Researchers at Michigan Medicine have uncovered the molecular mechanism that drives the disease-causing effects of the most common genetic risk factor for lupus, a study suggests.
Systemic lupus erythematosus is a common, incurable autoimmune disease that affects millions of individuals worldwide, with a particularly high prevalence among women. A genetic variant, called HLA-DRB1*03:01, is the greatest risk factor for the condition, which involves inflammation in many vital organs, and can lead to severe disability and death.
In a study recently published in Communications Biology, investigators found that a protein coded by that HLA variant triggers a cascade of molecular and cellular effects that can cause the inflammatory symptoms seen in lupus patients.
“For the first time, we have found the enigmatic mechanism that genetically predisposes people to the worst effects of the most typical form of lupus,” said Joseph Holoshitz, M.D., senior author of the paper and professor of internal medicine and rheumatology at University of Michigan Medical School. “The findings could potentially facilitate the discovery of safe, simple and effective treatments for SLE by targeting this new pathway.”
The results support a novel theory how genetic variants of the kind of HLA-DRB1*03:01 can lead to autoimmune diseases independent of antigen presentation, the traditionally studied mechanism, which has been long proposed but, so far, not directly proven.
“We have identified a chain of events in cell culture, as well as a mouse model of the disease, that demonstrate how the abnormalities that can cause lupus develop from the first effect of the risk gene, to signaling, all the way to immune abnormalities and clinical manifestations of lupus,” said Bruna Miglioranza Scavuzzi, Ph.D., first author of the paper and a postdoctoral research fellow in the Division of Rheumatology at the University of Michigan Medical School.
The findings of this study are reminiscent of previous findings in rheumatoid arthritis, another HLA-associated disease, that have paved the way for the development of small molecules to effectively treat arthritis in mice, Holoshitz says.
“Human trials in RA with those compounds are being carried out, and I hope that our novel findings will lead to similar efforts to ease the burden of millions of lupus patients as well,” he said.

Leave a Reply