by Rockefeller University
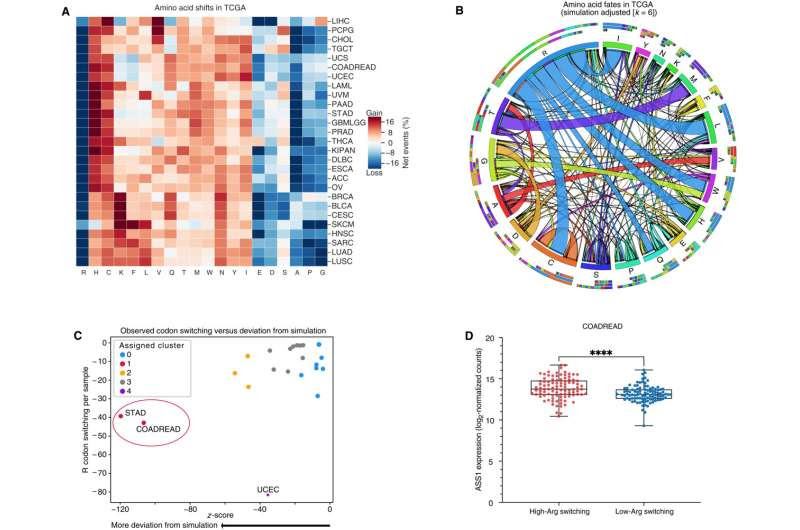
Arginine codons and residues are frequently lost and are associated with an increase in ASS1 expression. (A) Heatmap depicting codons gained (red) and lost (blue) across the TCGA. Gains and losses are normalized to the total number of missense and silent mutation events per sample for each cancer type. (B) Qualitative chord diagram showing amino acid switching events in cancer after adjustment from simulations. Ribbons that directly touch a column segment indicate loss of that specific amino acid codon during a mutational event and gain of the corresponding amino acid codon in which the ribbon terminates. Ribbons that begin and end at the same amino acid represent synonymous mutations. (C) Arginine codon-switching events observed versus predicted. Clusters were assigned with Affinity Propagation. (D) ASS1 expression in colorectal cancer (CRC) samples with either a high-degree or low-degree of arginine codon–switching events (n = 96 per group) with whiskers denoting minimum and maximum values. (DESeq2, ****P adjusted is less than 0.0001). Credit: Science Advances (2023). DOI: 10.1126/sciadv.ade9120
Arginine is an amino acid naturally produced by our bodies and plentiful in the fish, meat, and nuts that we eat. But as recent research in Science Advances reveals, arginine is an essential nutrient for cancer cells too. And starving them of it could potentially render tumors more vulnerable to the body’s natural immune response.
Researchers from Sohail Tavazoie’s Laboratory of Systems Cancer Biology at Rockefeller University have found that in a variety of human cancers, this amino acid becomes limited, prompting these cells to seek a clever genetic workaround: When arginine levels drop, they manipulate proteins at their disposal to more efficiently take up arginine and other amino acids. And remarkably, in a bid to keep growing, they induce mutations that reduce their reliance on it.
“It’s like if you had a Lego set, and you’re trying to build a fancy model plane, and you run out of the right bricks,” says first author Dennis Hsu, a former member of Tavazoie’s lab and now a physician-scientist at UPMC Hillman Cancer Center in Pittsburgh. “The only way to still build the plane would be if you had altered blueprints that don’t require the missing bricks.”
The arginine-cancer connection
On a cellular level, arginine plays a role in a variety of processes, from nitrogen waste disposal to protein synthesis. It’s also one the few amino acids that has been shown to regulate how immune cells react to cancer and other sorts of immunologic triggers, Hsu says.
Its deficit, for instance, is linked to the inflamed tissues of people with Crohn’s disease, ulcerative colitis, inflammatory bowel disease, or an H. pylori infection, whose tissues can have low levels of arginine. If people with these conditions do not get treated, they have a higher risk of developing stomach or colon cancer.
The researchers uncovered the arginine-cancer connection as part of a larger study on codons, triplets of DNA bases that each contain the recipe for producing a single amino acid. In combing through the Cancer Genome Atlas, Hsu documented thousands of instances of codon mutations, but one stood out among all cancers: arginine codons, which were lost during mutations far more than they should’ve been. Stomach and colorectal cancers showed the most dramatic deficiency.
“This was a very surprising discovery that we were not expecting,” says Tavazoie.
The researchers don’t know how the initial arginine drops came about. “We think that some cancers develop under low-arginine conditions and carry this history in their DNA,” says Hsu.
Cellular malnutrition
Hsu and his co-authors spent months in the lab growing cancer cells, and then starving them of arginine. As they put the cells through multiple rounds of cellular malnutrition, the cancer cells began mutating as they tried different ways to secure access to a renewed supply of the dietary essential. Not all of these strategies worked.
One successful method was increasing the amount of amino acid transporter proteins so that the cells could more efficiently take up arginine and other amino acids. But even these functional knockoffs tended to be meager sources. Meanwhile, the errors compounded as the cells replicated, resulting in changes to the genome such as mutated genes and misshapen proteins.
In another experiment, Hsu found an increase in the number of mutations towards codons that produce amino acids that were more abundant in the environment of the cancer cells. These suddenly became more appetizing to the cancer cells, which seemed to be trying to make do with what they had—akin to cobbling together a meal out of a few random items that happen to be in your fridge.
Linking a specific nutrient to a specific DNA change through this sort of so-called directed evolution “had not been reported before, to our knowledge,” Tavazoie says.
From red flag to bullseye
Interestingly, this ability to coax codons into doing their bidding could potentially lead to the cancer cells’ undoing. That’s because in the process of trying to keep themselves alive while malnourished, the cells accumulate so many mutations that they may begin to look very strange to the immune system.
“You have a bunch of random, abnormal-looking proteins because of all the mutations, and those are more likely to be recognized by the immune system as something that shouldn’t belong there,” Hsu says. Once deeply mutated, arginine-starved cancer cells that might’ve been able to fly under the radar of the immune system might now be waving a tattered red flag at it.
The findings have potential implications for immunotherapy. “By starving a cancer cell, perhaps you can promote the gain of new mutations that can then be recognized by the immune system,” Hsu says. “We have not tested this, but it would be a really cool thing to try.”

Leave a Reply