BIOTECHNOLOGY AND HEALTH
Just like real atlases, brain atlases vary widely in scale and content.
By Cassandra Willyardarchive page
May 10, 2024
“”
This rendering shows all of the excitatory (pyramidal) neurons in a part of the brain sample, at varying degrees of magnification and tilt. They are colored by size; the cell body (central core) of the cells ranges from 15-30 micrometers across.
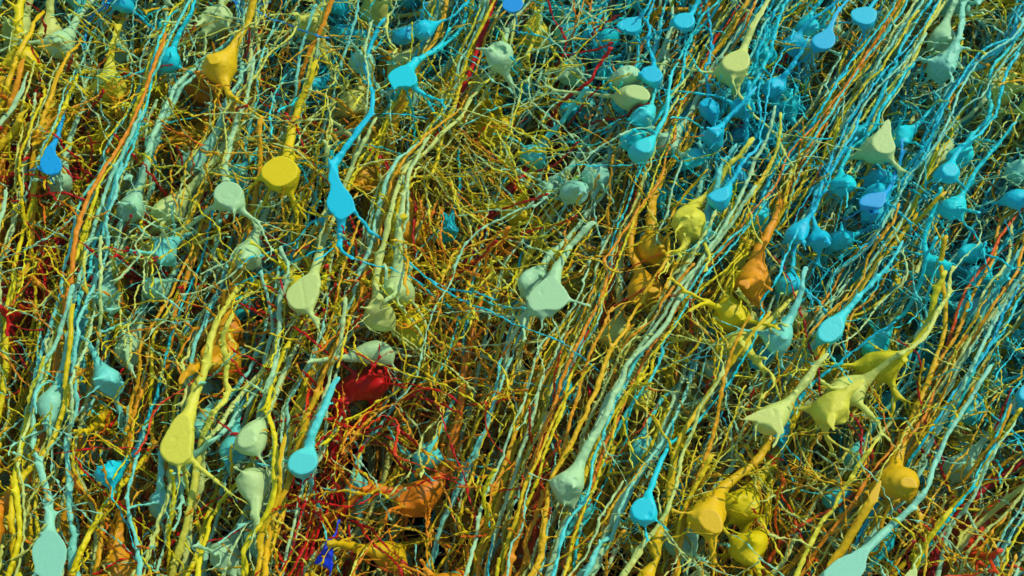
The human brain is an engineering marvel: 86 billion neurons form some 100 trillion connections to create a network so complex that it is, ironically, mind boggling.
This week scientists published the highest-resolution map yet of one small piece of the brain, a tissue sample one cubic millimeter in size. The resulting data set comprised 1,400 terabytes. (If they were to reconstruct the entire human brain, the data set would be a full zettabyte. That’s a billion terabytes. That’s roughly a year’s worth of all the digital content in the world.)
This map is just one of many that have been in the news in recent years. (I wrote about another brain map last year.) So this week I thought we could walk through some of the ways researchers make these maps and how they hope to use them.
Scientists have been trying to map the brain for as long as they’ve been studying it. One of the most well-known brain maps came from German anatomist Korbinian Brodmann. In the early 1900s, he took sections of the brain that had been stained to highlight their structure and drew maps by hand, with 52 different areas divided according to how the neurons were organized. “He conjectured that they must do different things because the structure of their staining patterns are different,” says Michael Hawrylycz, a computational neuroscientist at the Allen Institute for Brain Science. Updated versions of his maps are still used today.
“With modern technology, we’ve been able to bring a lot more power to the construction,” he says. And over the past couple of decades we’ve seen an explosion of large, richly funded mapping efforts.
BigBrain, which was released in 2013, is a 3D rendering of the brain of a single donor, a 65-year-old woman. To create the atlas, researchers sliced the brain into more than 7,000 sections, took detailed images of each one, and stitched the sections into a three-dimensional reconstruction.
In the Human Connectome Project, researchers scanned 1,200 volunteers in MRI machines to map structural and functional connections in the brain. “They were able to map out what regions were activated in the brain at different times under different activities,” Hawrylycz says.
A massive suite of papers offers a high-res view of the human and non-human primate brain.
This kind of noninvasive imaging can provide valuable data, but “Its resolution is extremely coarse,” he adds. “Voxels [think: a 3D pixel] are of the size of a millimeter to three millimeters.”
And there are other projects too. The Synchrotron for Neuroscience—an Asia Pacific Strategic Enterprise, a.k.a. “SYNAPSE,” aims to map the connections of an entire human brain at a very fine-grain resolution using synchrotron x-ray microscopy. The EBRAINS human brain atlas contains information on anatomy, connectivity, and function.
The work I wrote about last year is part of the $3 billion federally funded Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative, which launched in 2013. In this project, led by the Allen Institute for Brain Science, which has developed a number of brain atlases, researchers are working to develop a parts list detailing the vast array of cells in the human brain by sequencing single cells to look at gene expression. So far they’ve identified more than 3,000 types of brain cells, and they expect to find many more as they map more of the brain.
The draft map was based on brain tissue from just two donors. In the coming years, the team will add samples from hundreds more.
Mapping the cell types present in the brain seems like a straightforward task, but it’s not. The first stumbling block is deciding how to define a cell type. Seth Ament, a neuroscientist at the University of Maryland, likes to give his neuroscience graduate students a rundown of all the different ways brain cells can be defined: by their morphology, or by the way the cells fire, or by their activity during certain behaviors. But gene expression may be the Rosetta stone brain researchers have been looking for, he says: “If you look at cells from the perspective of just what genes are turned on in them, it corresponds almost one to one to all of those other kinds of properties of cells.” That’s the most remarkable discovery from all the cell atlases, he adds.
I have always assumed the point of all these atlases is to gain a better understanding of the brain. But Jeff Lichtman, a neuroscientist at Harvard University, doesn’t think “understanding” is the right word. He likens trying to understand the human brain to trying to understand New York City. It’s impossible. “There’s millions of things going on simultaneously, and everything is working, interacting, in different ways,” he says. “It’s too complicated.”
But as this latest paper shows, it is possible to describe the human brain in excruciating detail. “Having a satisfactory description means simply that if I look at a brain, I’m no longer surprised,” Lichtman says. That day is a long way off, though. The data Lichtman and his colleagues published this week was full of surprises—and many more are waiting to be uncovered.

Leave a Reply