Diana Kwon
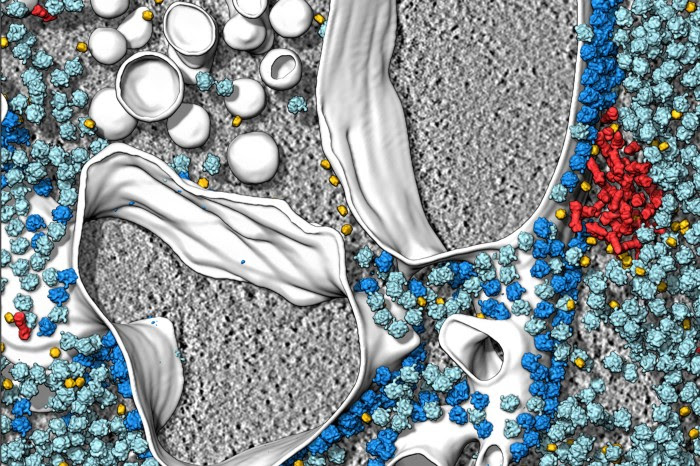
Cryo-electron tomography and related techniques can showcase the insides of cells in striking detail. Credit: S. Albert et al./PNAS (CC BY 4.0)
For a few weeks in 2017, Wanda Kukulski found herself binge-watching an unusual kind of film: videos of the insides of cells. They were made using a technique called cryo-electron tomography (cryo-ET) that allows researchers to view the proteins in cells at high resolution. In these videos, she could see all kinds of striking things, such as the inner workings of cells and the compartments inside them, in unprecedented detail. “I was so overwhelmed by the beauty and the complexity that in the evenings I would just watch them like I would watch a documentary,” recalls Kukulski, a biochemist at the University of Bern, Switzerland.
In recent years, imaging techniques such as cryo-ET have started to enable scientists to see biological molecules in their native environments. Unlike older methods that take individual proteins out of their niches to study them, these techniques provide a holistic view of proteins and other molecules together with the cellular landscape. Although they still have limitations — some researchers say that the resolution of cryo-ET, for example, is too low for molecules to be identified with certainty — the techniques are increasing in popularity and sophistication. Researchers who turn to them are not only mesmerized by the beautiful images, but also blown away by some of the secrets that are being revealed — such as the tricks bacteria use to infect cells or how mutated proteins drive neurodegenerative diseases such as Parkinson’s.
Every peek through the microscope is another chance to explore uncharted cellular terrain, says Grant Jensen, a structural biologist at the California Institute of Technology in Pasadena. “There’s definitely a great joy in being able to see something for the first time,” he says.
Other researchers share his delight. Elizabeth Villa, a biophysicist at the University of California, San Diego, recalls her overwhelming excitement the first time she saw cell structures with cryo-ET. “It felt as if all of a sudden, we were paparazzi with access we never had before,” Villa says.
From crystals to context
For decades, researchers have relied on a technique called X-ray crystallography to visualize proteins, viruses, and other biological entities. The method involves coaxing molecules into forming static, well-ordered crystals and then bombarding the samples with intense X-ray beams. It enabled the discovery of the helical nature of DNA and the structure of more than 100,000 proteins, but it has its limitations: crystallizing molecules is difficult and tedious, and not always possible.
Scientists have overcome these drawbacks using cryo-electron microscopy (cryo-EM), a technique that reveals the structure of biomolecules that have been isolated from their surroundings and then frozen. In cryo-EM, the samples are showered with beams of electrons. Although the technique was initially ridiculed as ‘blobology’, owing to the blurry images it produced, advances in sample preparation and image processing have increased its resolution enough to visualize individual atoms (around 1.2 ångströms or 1.2 × 10–10 m in size).
As this ‘resolution revolution’ started to sweep through cryo-EM, around 2013, scientists flocked to the method. So far, researchers have used it to solve the structures of more than 10,000 biological molecules. Proteins found in cell membranes, in particular, have been of interest, because many of them are important for understanding disease and developing drugs. The advance “opened the door for some of those really talented people to then look for the next richest, most ripe field for big impact advances”, says Jensen. That area happened to be cryo-ET.
Its early proponents sought a technique that could view biological molecules not only in fine detail, but also as they would look inside cells. Like cryo-EM, cryo-ET requires an electron microscope and relies on a sample preparation method known as vitrification: the rapid cooling of the water around a sample so that it freezes into a glass-like state, rather than as ice crystals. Unlike conventional cryo-EM, however, which requires purified samples, investigators can use cryo-ET to capture these molecules in situ.
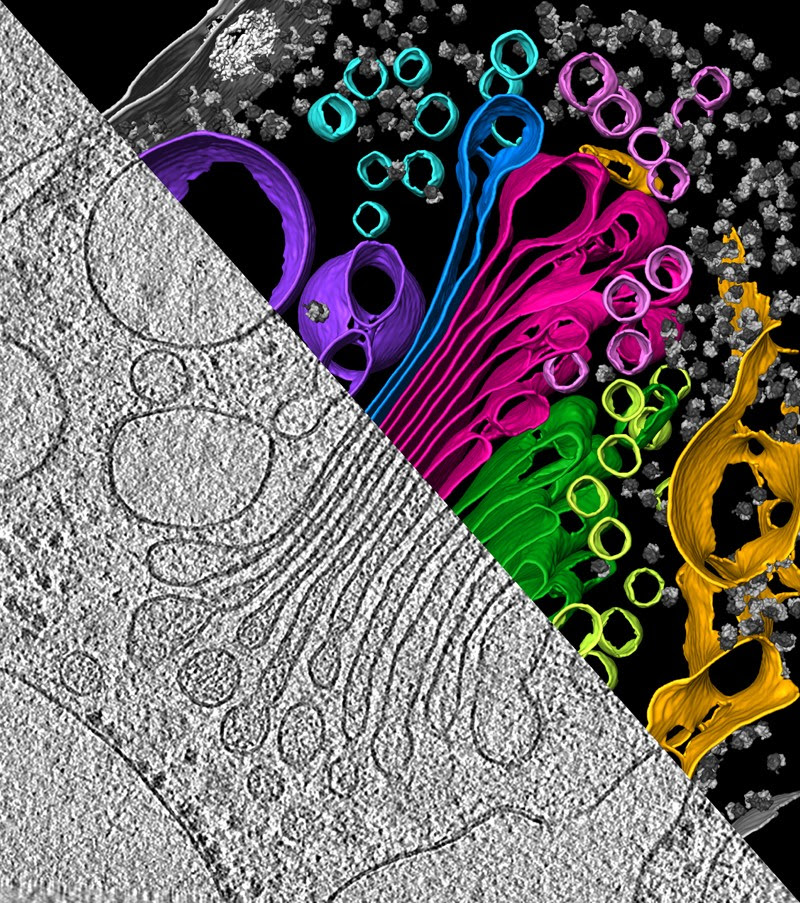
An algal cell reveals its secrets under cryo-ET, showing the endoplasmic reticulum (yellow), which makes proteins, the pouches of the Golgi apparatus (green and magenta), which modifies and packages proteins for transport, and vesicles (small circles, various colours), which carry proteins.Credit: Y. S. Bykkov et al./eLIFE (CC BY 4.0)
With cryo-EM, scientists make a 3D image by taking 2D pictures of lots of isolated molecules in different configurations, and merging the results. With cryo-ET, by contrast, they take multiple snapshots of a single chunk of material, teeming with molecules, from many different angles, allowing the surroundings to be kept intact.
It’s like having a photo of a whole crowd, rather than one person’s headshot. This is why Wolfgang Baumeister, a biophysicist at the Max Planck Institute of Biochemistry in Martinsried, Germany, who is one of the pioneers of the technique, and his colleagues have dubbed it “molecular sociology”.
And this is how proteins live, after all. “Proteins are social — at any given time a protein is in a complex with about ten other proteins,” says Villa. After viewing such interactions with cryo-ET, “I could not stomach the thought of myself studying another protein in isolation,” she adds.
Electron tomography itself — the use of an electron microscope to image a specimen from several angles — has been around since the 1960s, but it wasn’t until the 1990s that the method began to come into its own. One of the challenges was that streams of electrons are extremely damaging to biological samples, which made it difficult to capture enough snapshots for a clear, crisp image. Scientists have sharpened their pictures using the latest sample-slicing processes and computational methods. For instance, a technique called cryo-focused-ion-beam (cryo-FIB) milling can cut samples into extremely thin slices known as lamellae. Still, the cost and technical expertise needed to use cryo-ET — particularly in combination with methods such as cryo-FIB milling — can be prohibitive for many laboratories, Baumeister says.
Early demonstrations of cryo-ET from Baumeister’s group included snapshots1 of the cells of Dictyostelium, a bacteria-guzzling amoeba that lives in soil. The team revealed, among other things, previously unseen characteristics of intricate protein networks that make up the amoeba’s cytoskeleton — such as how individual filaments interact with one another and attach to specific structures on the membranes of Dictyostelium cells.
”You can rarely assign biological functions or cellular functions to an individual molecule — the functions arise from the interaction of all the molecules inhabiting a cellular landscape,” Baumeister says. “That’s where the discovery potential of cryo-ET comes in. Whatever we look at nowadays, there are surprises.”
Sociable cells
Much of the early work with cryo-ET was on prokaryotes: single-celled organisms such as bacteria. These cells are usually smaller and thinner than the more complex cells of eukaryotes.
In a study of Jensen’s from 2006, for example, he and his team reported the first complete structure of the motor that drives the flagellum, a whip-like appendage in bacteria2. Using cryo-ET, they unveiled the architecture of the 20-piece motor in Treponema primitia, a bacterium found in termites’ guts, and detailed how these parts were positioned in the bacterial membrane. Jensen and his colleagues have also revealed key details of bacterial pili — protrusions that the microorganisms use for many functions, such as attaching to and secreting substances into cells they infect. Last year, his team released an open-access, digital atlas (see go.nature.com/3nugs7v) highlighting the many insights cryo-ET has revealed about bacterial and other prokaryotic cells.
More recently, scientists have moved on to imaging eukaryotic cells — which are palatial in comparison with prokaryotes. This has been possible largely because of the advent of cryo-FIB milling, which allows researchers to slice cells thinly before placing them under an electron microscope. Baumeister and his colleagues used this mash-up of methods to visualize how molecules were arranged in the vicinity of the nucleus in a human cell3 (see ‘Inside scoop’). Their work revealed how previously unseen, nanometre-thin filaments provided structural support to the nucleus — making it one of the stiffest organelles in animal cells.
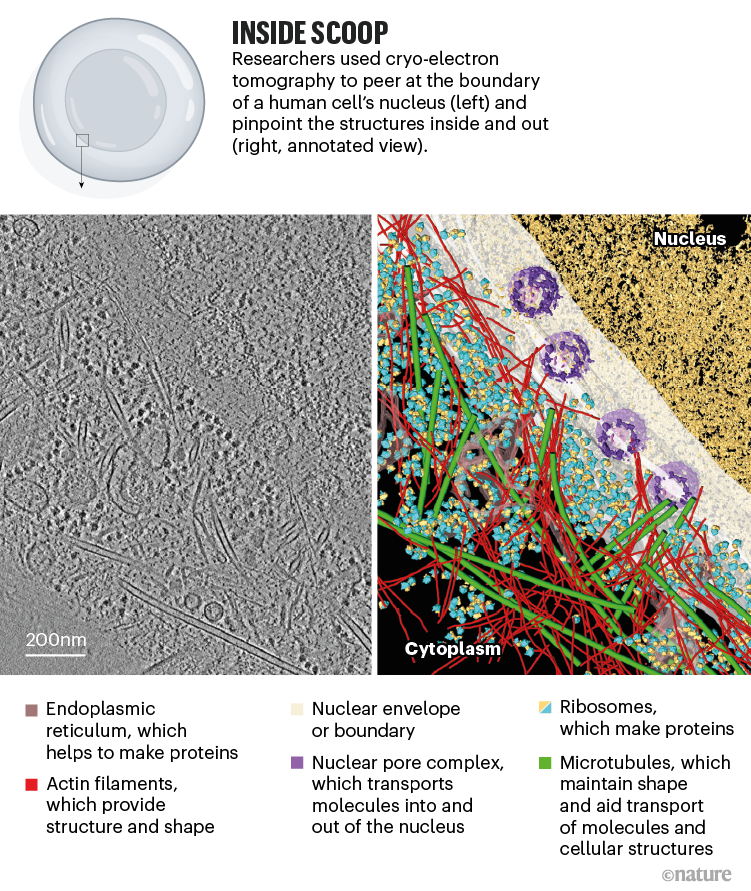
Source: Ref. 3
Even with FIB milling, cryo-ET can capture only tiny segments of eukaryotic cells, meaning that scientists need to find a way to pinpoint molecules of interest in a vast and crowded cellular landscape. One solution is to pick out proteins by first fluorescently labelling them under a light microscope, and then zooming into the finer details in specific sections using cryo-ET.
Villa and her colleagues have used this combination of techniques to resolve the architecture of LRRK2, a protein linked to inherited forms of Parkinson’s disease4. Their work revealed that the mutated version of the protein stuck to components of the cytoskeleton known as microtubules, forming a double helix around them. The team’s findings also hinted that mutated LRRK2 might form a configuration that promotes this type of binding — potentially causing problems by blocking molecules that carry important cellular cargo along the microtubules5.
Groups such as Baumeister’s have used this method to examine how proteins associated with neurogenerative diseases such as Huntington’s6 and motor neuron disease (amyotrophic lateral sclerosis, or ALS)7 interact with components of the cell such as the endoplasmic reticulum (ER), a large piece of cellular machinery that helps to synthesize proteins. Researchers have found that neurotoxic clumps of proteins implicated in these diseases behave very differently to one another inside cells. In Huntington’s, for example, aggregates of a mutant form of a protein called huntingtin seem to throw the organization of the ER into disarray, whereas in ALS, aggregates of an abnormal protein impair the cell’s biochemistry by activating its protein-degrading machinery.
In the future, scientists hope to use such methods to better understand how therapeutics work, by visualizing how drugs act on the molecular innards of cells. In an early demonstration, Julia Mahamid at the European Molecular Biology Laboratory in Heidelberg, Germany, and her colleagues were able to glimpse antibiotics in a bacterial cell binding to ribosomes — organelles that serve as protein factories8. The feat was made possible by pushing the resolution of cryo-ET to 3.5 ångströms.
“I think that’s probably the state of the art of what’s possible [with cryo-ET],” says Kukulski, who was not involved in this work. However, she notes that the ribosome is ubiquitous in the cell and is already well characterized, making it easy to recognize and study. Trying to image lesser-known or rare cellular structures remains an incredibly difficult task, she adds.
Powerful combinations
Cryo-ET is a fast-growing field, but the technique still has a number of limitations. Resolution remains an issue. Although the level of detail has improved drastically in recent years, cryo-ET is unable to attain the atomic-level resolution of cryo-EM. “Cryo-ET is where cryo-EM was in the early 90s, long before it was able to achieve atomic resolution,” says Hong Zhou, a biophysicist at the University of California, Los Angeles.
At the current level of performance, it is difficult to correctly identify molecules in a cell using cryo-ET, according to Zhou. So, unless scientists are looking at structures that have previously been well characterized, such as the ribosome, their hypotheses about what they see with the technique might prove to be incorrect, he adds. “The odds are against you. It’s very easy to make mistakes.”
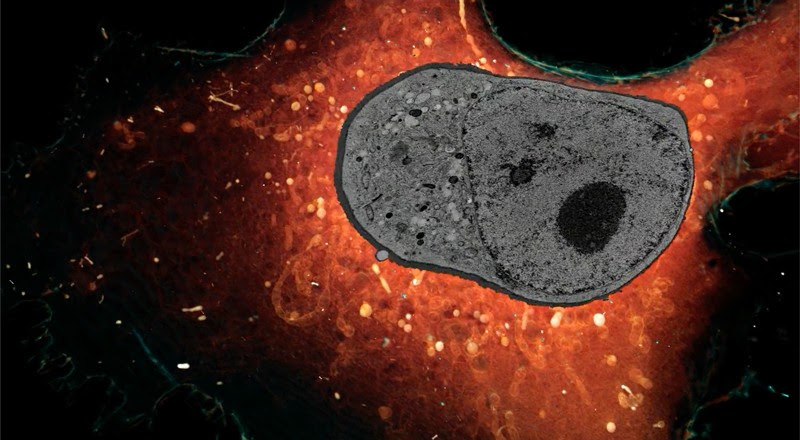
Whole cells sliced very thinly and imaged with super-resolution microscopy. Credit: Janelia Research Campus HHMI
To circumvent the issue of resolution, Zhou has chosen to try to push the limits of conventional cryo-EM instead. His team recently reported a method called cryoID9, which melds cryo-EM with various other techniques. One of these involves breaking open cells in a method that enables proteins to partially remain inside the original cellular milieu; in this way, researchers can view proteins in a near-native state. Despite his current focus on cryo-EM, Zhou says that cryo-ET is the future. “I consider [this method] an intermediate step towards that goal.”
Another limitation of cryo-ET is its narrow sampling. “The best-kept secret of tomography is that when you look at a mammalian cell, a tomograph is less than 0.1% of that cell,” Villa says. This means that large organelles such as the nucleus can be viewed only in tiny segments. To fill this gap, scientists such as Harald Hess, a biophysicist at the Howard Hughes Medical Institute’s Janelia Research Campus in Ashburn, Virginia, are using techniques other than cryo-ET, namely super-resolution fluorescence microscopy and electron microscopy, to visualize entire cells. Using these methods, Hess and his colleagues are gaining a fresh view into how various cellular components interact. In a study published earlier this month, the researchers demonstrated that by using machine learning — a form of artificial intelligence — to help identify these components in many samples, they could map the organization of up to 35 types of organelle1.
Other researchers are combining cryo-ET with another technique called X-ray tomography that can capture images of whole cells. This allows scientists to examine the structure of larger components, such as mitochondria or nuclei, and then zoom in on specific areas of interest.
However, bringing these methods together requires both money and skill. On top of that, both techniques bombard samples with damaging radiation. That makes it a challenge to transfer a sample between them, says Eva Pereiro, a beamline scientist at the ALBA synchrotron facility in Barcelona, Spain, that produces X-rays suitable for tomography.
Some labs have already accomplished this feat. Maria Harkiolaki, principal beam-line scientist at the Diamond Light Source, a synchrotron facility in Didcot, UK, and her colleagues recently published2 a model of the mechanics of SARS-CoV-2 infection that uses cryo-ET and X-ray tomography to elucidate the process. They captured events at both the level of the cell and individual molecules, and proposed an idea for how the virus replicates in primate cells.
Baumeister thinks that, like cryo-EM, cryo-ET will eventually allow scientists to view biological molecules in atomic detail. Until then, scientists continue to eagerly investigate what insights into the cell might be revealed by cryo-ET and other similar methods. Because these tools can reveal structures that have never been seen before, researchers are often left with new mysteries to solve. “What I love about tomography”, Villa says, “is that we always generate more questions than answers.”

Leave a Reply