by Wellcome Trust Sanger Institute
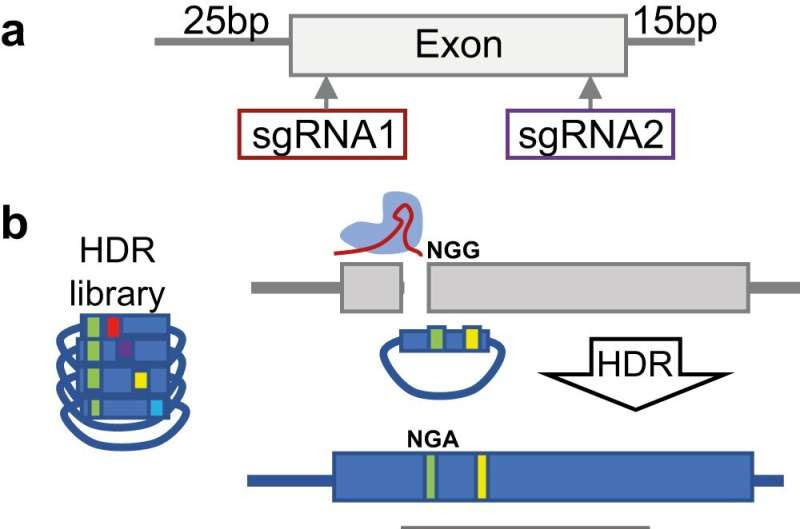
Experimental design overview. a Two independent sgRNAs and associated HDR variant libraries are designed at the 5′ and 3′ end of each exon. b The sgRNA, together with the HDR template library are transfected into LIG4-KO Cas9-expressing HAP1 cells. HDR utilizes the library as a template for repair of the sgRNA-directed double-stranded DNA cut, incorporating a DDX3X variant of interest. Damaging DDX3X variants will reduce cell viability or proliferation. Variant abundance was assessed at five timepoints. Functional missense (purple) and synonymous variants (Syn, blue) remain abundant, while loss-of-function variants (LOF, red), and damaging missense (yellow) variants are depleted. Credit: Nature Communications (2023). DOI: 10.1038/s41467-023-43041-4
Researchers now understand the functional impact of thousands of genetic changes within the DDX3X gene. This could lead to enhanced diagnosis and treatment of various neurodevelopmental disorders and cancers.
Their new tool outperforms rivals in determining the genetic basis of neurodevelopmental disorders and cancer, identifying harmful mutations at scale, previously too complex to interpret. It promises new avenues for diagnosis and treatment.
Researchers from the Wellcome Sanger Institute and their collaborators at the University of Cambridge investigated and validated the functional impact of more than 12,000 genetic changes within the DDX3X gene. They found around a quarter of the genetic changes within DDX3X negatively affect its function, revealing the importance of 90% of previously unexplained genetic changes’ impact on health.
Comparing this finding with patient data, the researchers confirm this loss of DDX3X function is a significant contributor to neurodevelopmental disorders and is a key player in cancer development.
The findings, published today (6 December) in Nature Communications, shed light on previously inscrutable aspects of the human genetic code, helping provide valuable insights into the genetic mechanisms underlying neurodevelopmental disorders and cancers. The work paves the way for new early detection methods and treatments.
It is hoped the new technique can be broadly applied to understand how changes in many other genes are relevant for neurodevelopmental disorders.
The DDX3X gene has long been associated with neurodevelopmental disorders, particularly in women, and certain forms of cancer. These disorders are typically associated with intellectual disabilities, developmental delays, and often features such as seizures, movement disorders, and behavioral problems.
Diagnosis is highly challenging, especially in young children with unclear symptoms or unborn babies, leading to misdiagnoses of other conditions like autism. Detecting these neurodevelopmental disorders early through genetic screening can greatly enhance treatment effectiveness and improve quality of life for individuals affected, but until now there has been limited understanding of which harmful genetic changes to look out for.
In this new study, scientists set out to uncover the impact of all possible genetic changes within the DDX3X gene on protein function and health, including neurodevelopmental disorders and cancer.
Unlike computer-based predictive tools, the team integrated real experimentation to test thousands of these genetic changes by artificially altering the genetic code of human cells grown in a dish, in a process known as “saturation genome editing.” To understand the effects of having these genetic alterations, they compared the experimental data with health data from the UK Biobank cohort.
The team identified that 3,432 of the 12,776 different genetic changes had a negative impact on the protein’s function. Using the technique, they were able to shed light on the significance of up to 93% of genetic changes, for which the impact on health was previously unknown. They were able to achieve an accuracy rate of at least 97% in pinpointing DDX3X genetic changes related to neurodevelopmental disorders.
Researchers also found that genetic changes seen in cancer prevent the DDX3X protein from working properly, carrying implications for the development of new cancer treatments targeting the gene.
Together, these findings advance the understanding of mechanisms underlying neurodevelopmental disorders and cancer. They provide clinicians with valuable insights into the potential impact of genetic changes in DDX3X on a child’s health, aiding in earlier diagnosis.
“In the context of genetic conditions, even minor changes in the genetic code can have profound implications for a child’s development. Our approach, which goes beyond computation to assess protein function, overcomes this diagnostic challenge to reliably distinguish between harmless and harmful rare genetic changes. We hope to apply this technique to other genes, unlocking essential insights hidden within our genetic code,” says Dr. Sebastian Gerety.
“DDX3X is altered in a range of cancers and in particular in childhood brain cancers. Understanding exactly which mutations are disease-causing facilitates diagnosis and can help ensure patients get the most suitable treatment for their disease,” says Dr. David Adams.
“Genetic testing is increasingly integrated into patient care, yet our ability to decode the genetic information has not kept pace, preventing families from receiving the full support they need. These freely available insights will empower doctors to interpret genetic tests and diagnose children earlier, enabling timely intervention and improved quality of life for those affected by DDX3X-linked neurodevelopmental disorders,” says Dr. Elizabeth Radford.
More information: Elizabeth J. Radford et al, Saturation genome editing of DDX3X clarifies pathogenicity of germline and somatic variation, Nature Communications (2023). DOI: 10.1038/s41467-023-43041-4
Journal information: Nature Communications
Provided by Wellcome Trust Sanger Institute

Leave a Reply